Tutorial: Profiling a PyTorch Model#
In this tutorial, we will use Metinor to profile a PyTorch model. Profiling a model is important to understand the performance of the model and to identify bottlenecks. Metinor provides a simple API to profile a model and a set of tools to analyze and plot the results.
[1]:
# Suppress warnings (to make the output cleaner in this notebook)
import warnings
warnings.filterwarnings("ignore")
Create a model#
The profiler in Metinor supports subclasses of nn.Module. In this tutorial, we will use a pre-trained ResNet-18 from torchvision.
[2]:
from torchvision.models import resnet18, ResNet18_Weights
# Create the model to profile
model = resnet18(weights=ResNet18_Weights.DEFAULT)
input_shape = (3, 224, 224)
Import the profiler#
Next, we will import the profiler from the metinor.profiler package. This is available as metinor.profiler.Profiler class and several metinor.functional.profiler.profile_* functions. We will profile both the model types using the profiler class and the functions.
Profiling a model using the functional API#
[3]:
from nervai_optim.functional.profiler import profile_metrics
# Profile the model
model_report = profile_metrics(model, input_shape=input_shape)
# Report is a `metinor.profiler.ProfileReport` object which contains the following variables:
# - `report.summary`: A summary of the profiled model
# - `report.details`: A layer-by-layer breakdown of the profiled model
# - `report.pretty_summary`: A human-readable summary of the profiled model
# - `report.pretty_details`: A human-readable layer-by-layer breakdown of the profiled model
# Each of these variables is a `pandas.DataFrame` object
# Print the summary of the profiled model
model_report.pretty_summary
[3]:
| ProfilerMetric.NAME | ProfilerMetric.TYPE | ProfilerMetric.INPUTS | ProfilerMetric.OUTPUTS | ProfilerMetric.PARAMS | ProfilerMetric.PARAMS_TRAINABLE | ProfilerMetric.FLOPS | ProfilerMetric.MADDS | ProfilerMetric.MEM_READ | ProfilerMetric.MEM_WRITE | ProfilerMetric.RUNTIME | ProfilerMetric.RMS | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | ResNet | ResNet | (3, 224, 224) | (1000,) | 11.69M | 11.69M | 1.82G | 3.64G | 18.71M | 6.72M | 0.028 | 7.07 |
[4]:
# Show some details of first 10 layers
from nervai_optim.profiler.metrics import ProfilerMetric
columns = [
ProfilerMetric.NAME,
ProfilerMetric.TYPE,
ProfilerMetric.FLOPS,
ProfilerMetric.MADDS,
ProfilerMetric.RUNTIME,
]
model_report.pretty_details[columns].head(10)
[4]:
| ProfilerMetric.NAME | ProfilerMetric.TYPE | ProfilerMetric.FLOPS | ProfilerMetric.MADDS | ProfilerMetric.RUNTIME | |
|---|---|---|---|---|---|
| 1 | conv1 | Conv2d | 118.01M | 235.23M | 0.0025 |
| 2 | bn1 | BatchNorm2d | 1.61M | 3.21M | 0.0006 |
| 3 | relu | ReLU | 802.82K | 802.82K | 0.00014 |
| 4 | maxpool | MaxPool2d | 802.82K | 1.61M | 0.0013 |
| 5 | layer1.0.conv1 | Conv2d | 115.61M | 231.01M | 0.0012 |
| 6 | layer1.0.bn1 | BatchNorm2d | 401.41K | 802.82K | 0.00022 |
| 7 | layer1.0.relu | ReLU | 200.7K | 200.7K | 1.7881393432617188e-05 |
| 8 | layer1.0.conv2 | Conv2d | 115.61M | 231.01M | 0.0012 |
| 9 | layer1.0.bn2 | BatchNorm2d | 401.41K | 802.82K | 0.00019 |
| 10 | layer1.1.conv1 | Conv2d | 115.61M | 231.01M | 0.0008 |
[5]:
# More functional API examples
from nervai_optim.functional.profiler import (
profile_flops,
profile_runtime,
profile_memory,
profile_madds,
profile_params,
)
# Profile the model
params = profile_params(model, input_shape=input_shape, readable=True)
flops = profile_flops(model, input_shape=input_shape, readable=True)
runtime = profile_runtime(model, input_shape=input_shape, readable=True)
memory = profile_memory(model, input_shape=input_shape, readable=True)
madds = profile_madds(model, input_shape=input_shape, readable=True)
print("ResNet-18 Profiling Results")
print("-------")
print(f"Params: {params}")
print(f"FLOPs: {flops}")
print(f"MAdds: {madds}")
print(f"Memory: {memory}")
print(f"Runtime: {runtime}")
ResNet-18 Profiling Results
-------
Params: 11.69M
FLOPs: 1.82G
MAdds: 3.64G
Memory: ('18.71M', '6.72M')
Runtime: 0.028
Profiling a model using the profiler class#
Next, we will profile the model using the profiler class.
[6]:
from nervai_optim.profiler import Profiler, ProfilerReport
# Create a profiler object and profile the model
profiler = Profiler(model, input_shape=input_shape)
profiled_nodes = profiler.profile()
# `profiled_nodes` is a list of `metinor.profiler.ProfiledNode` objects, which
# can be used to generate a report using `metinor.profiler.ProfilerReport` class
model_report = ProfilerReport(profiled_nodes)
model_report.pretty_summary
[6]:
| ProfilerMetric.NAME | ProfilerMetric.TYPE | ProfilerMetric.INPUTS | ProfilerMetric.OUTPUTS | ProfilerMetric.PARAMS | ProfilerMetric.PARAMS_TRAINABLE | ProfilerMetric.FLOPS | ProfilerMetric.MADDS | ProfilerMetric.MEM_READ | ProfilerMetric.MEM_WRITE | ProfilerMetric.RUNTIME | ProfilerMetric.RMS | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | ResNet | ResNet | (3, 224, 224) | (1000,) | 11.69M | 11.69M | 1.82G | 3.64G | 18.71M | 6.72M | 0.031 | 7.07 |
[7]:
model_report.pretty_details.head(15)
[7]:
| ProfilerMetric.NAME | ProfilerMetric.TYPE | ProfilerMetric.INPUTS | ProfilerMetric.OUTPUTS | ProfilerMetric.PARAMS | ProfilerMetric.PARAMS_TRAINABLE | ProfilerMetric.FLOPS | ProfilerMetric.MADDS | ProfilerMetric.MEM_READ | ProfilerMetric.MEM_WRITE | ProfilerMetric.RUNTIME | ProfilerMetric.RMS | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | conv1 | Conv2d | (3, 224, 224) | (64, 112, 112) | 9.41K | 9.41K | 118.01M | 235.23M | 159.94K | 802.82K | 0.0019 | 0.13 |
| 2 | bn1 | BatchNorm2d | (64, 112, 112) | (64, 112, 112) | 128 | 128 | 1.61M | 3.21M | 802.94K | 802.82K | 0.0008 | 0.32 |
| 3 | relu | ReLU | (64, 112, 112) | (64, 112, 112) | 0 | 0 | 802.82K | 802.82K | 802.82K | 802.82K | 0.00014 | 0.0 |
| 4 | maxpool | MaxPool2d | (64, 112, 112) | (64, 56, 56) | 0 | 0 | 802.82K | 1.61M | 802.82K | 200.7K | 0.0015 | 0.0 |
| 5 | layer1.0.conv1 | Conv2d | (64, 56, 56) | (64, 56, 56) | 36.86K | 36.86K | 115.61M | 231.01M | 237.57K | 200.7K | 0.0012 | 0.05 |
| 6 | layer1.0.bn1 | BatchNorm2d | (64, 56, 56) | (64, 56, 56) | 128 | 128 | 401.41K | 802.82K | 200.83K | 200.7K | 0.00017 | 0.3 |
| 7 | layer1.0.relu | ReLU | (64, 56, 56) | (64, 56, 56) | 0 | 0 | 200.7K | 200.7K | 200.7K | 200.7K | 1.9073486328125e-05 | 0.0 |
| 8 | layer1.0.conv2 | Conv2d | (64, 56, 56) | (64, 56, 56) | 36.86K | 36.86K | 115.61M | 231.01M | 237.57K | 200.7K | 0.001 | 0.045 |
| 9 | layer1.0.bn2 | BatchNorm2d | (64, 56, 56) | (64, 56, 56) | 128 | 128 | 401.41K | 802.82K | 200.83K | 200.7K | 0.00015 | 0.29 |
| 10 | layer1.1.conv1 | Conv2d | (64, 56, 56) | (64, 56, 56) | 36.86K | 36.86K | 115.61M | 231.01M | 237.57K | 200.7K | 0.0009 | 0.05 |
| 11 | layer1.1.bn1 | BatchNorm2d | (64, 56, 56) | (64, 56, 56) | 128 | 128 | 401.41K | 802.82K | 200.83K | 200.7K | 0.00028 | 0.27 |
| 12 | layer1.1.relu | ReLU | (64, 56, 56) | (64, 56, 56) | 0 | 0 | 200.7K | 200.7K | 200.7K | 200.7K | 1.9311904907226562e-05 | 0.0 |
| 13 | layer1.1.conv2 | Conv2d | (64, 56, 56) | (64, 56, 56) | 36.86K | 36.86K | 115.61M | 231.01M | 237.57K | 200.7K | 0.0011 | 0.044 |
| 14 | layer1.1.bn2 | BatchNorm2d | (64, 56, 56) | (64, 56, 56) | 128 | 128 | 401.41K | 802.82K | 200.83K | 200.7K | 0.00016 | 0.32 |
| 15 | layer2.0.conv1 | Conv2d | (64, 56, 56) | (128, 28, 28) | 73.73K | 73.73K | 57.8M | 115.51M | 274.43K | 100.35K | 0.0006 | 0.042 |
Analyzing the results#
In addition to the profiling results as a metinor.profiler.ProfilerReport object, Metinor also provides the metinor.profiler.analysis.ReportAnalyzer class to analyze the profiling results. This class provides several methods to search and filter the results. It also provides methods to plot the profiled metrics.
[8]:
from nervai_optim.profiler.analysis import ReportAnalyzer
# Create a report analyzer object
analyzer = ReportAnalyzer(model_report)
Find layers with the highest values of a metric#
The find_top_n_layers method can be used to find the top k layers based on a metric. It takes the metric name and the number of layers to return as arguments, and returns a pandas.DataFrame object with k highest values of the metric.
For example, in the code below, we will find the top 5 layers with the highest number of FLOPs.
[9]:
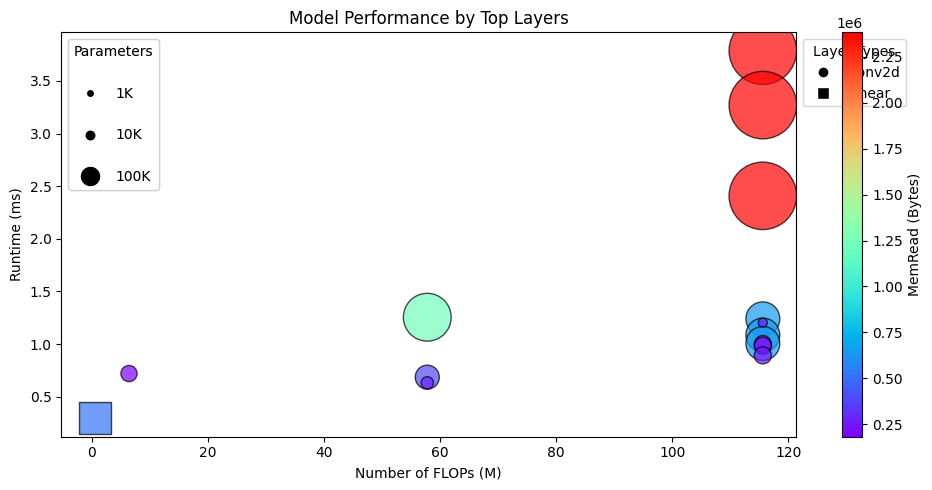
analyzer.find_top_n_layers(ProfilerMetric.PARAMS, n=5)
[9]:
| ProfilerMetric.NAME | ProfilerMetric.TYPE | ProfilerMetric.INPUTS | ProfilerMetric.OUTPUTS | ProfilerMetric.PARAMS | ProfilerMetric.PARAMS_TRAINABLE | ProfilerMetric.FLOPS | ProfilerMetric.MADDS | ProfilerMetric.MEM_READ | ProfilerMetric.MEM_WRITE | ProfilerMetric.RUNTIME | ProfilerMetric.RMS | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 42 | layer4.0.conv2 | Conv2d | (512, 7, 7) | (512, 7, 7) | 2359296 | 2359296 | 115605504 | 231185920 | 2384384 | 25088 | 0.003270 | tensor(0.0174) |
| 46 | layer4.1.conv1 | Conv2d | (512, 7, 7) | (512, 7, 7) | 2359296 | 2359296 | 115605504 | 231185920 | 2384384 | 25088 | 0.003787 | tensor(0.0180) |
| 49 | layer4.1.conv2 | Conv2d | (512, 7, 7) | (512, 7, 7) | 2359296 | 2359296 | 115605504 | 231185920 | 2384384 | 25088 | 0.002408 | tensor(0.0132) |
| 39 | layer4.0.conv1 | Conv2d | (256, 14, 14) | (512, 7, 7) | 1179648 | 1179648 | 57802752 | 115580416 | 1229824 | 25088 | 0.001255 | tensor(0.0199) |
| 30 | layer3.0.conv2 | Conv2d | (256, 14, 14) | (256, 14, 14) | 589824 | 589824 | 115605504 | 231160832 | 640000 | 50176 | 0.001086 | tensor(0.0251) |
The find_bottom_n_layers method does the same thing but returns the bottom k layers based on a metric.
Determine relative impact of each layer on a metric#
The layer_wise_impact method returns contribution of each layer to a specific metric. It takes the metric name as an argument and returns a pandas.DataFrame object with the value of that metric and a percentage contribution of each layer to that metric.
[10]:
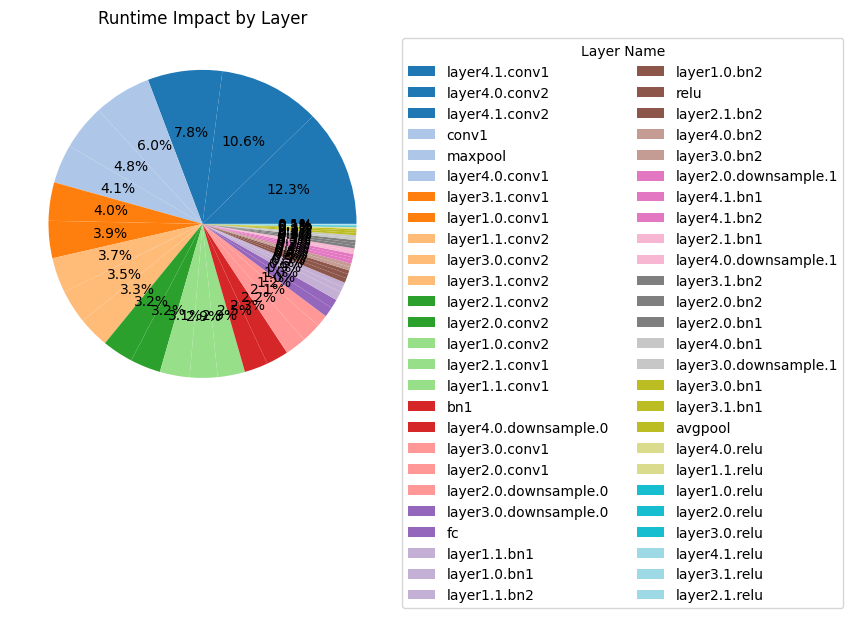
analyzer.layer_wise_impact(ProfilerMetric.RUNTIME).head(5)
[10]:
| ProfilerMetric.NAME | ProfilerMetric.TYPE | ProfilerMetric.RUNTIME | ProfilerMetric.RUNTIME (%) | |
|---|---|---|---|---|
| 46 | layer4.1.conv1 | Conv2d | 0.003787 | 12.306214 |
| 42 | layer4.0.conv2 | Conv2d | 0.003270 | 10.627319 |
| 49 | layer4.1.conv2 | Conv2d | 0.002408 | 7.825804 |
| 1 | conv1 | Conv2d | 0.001853 | 6.021399 |
| 4 | maxpool | MaxPool2d | 0.001475 | 4.791862 |
Filter layers by type#
The filter_by_layer_type method can be used to filter the layers by their type. It takes the layer type as an argument and returns a pandas.DataFrame object with the layers of the specified type. Note that all analyzer methods also take a keyword argument readable which can be set to True to return metrics in a human-readable format.
[11]:
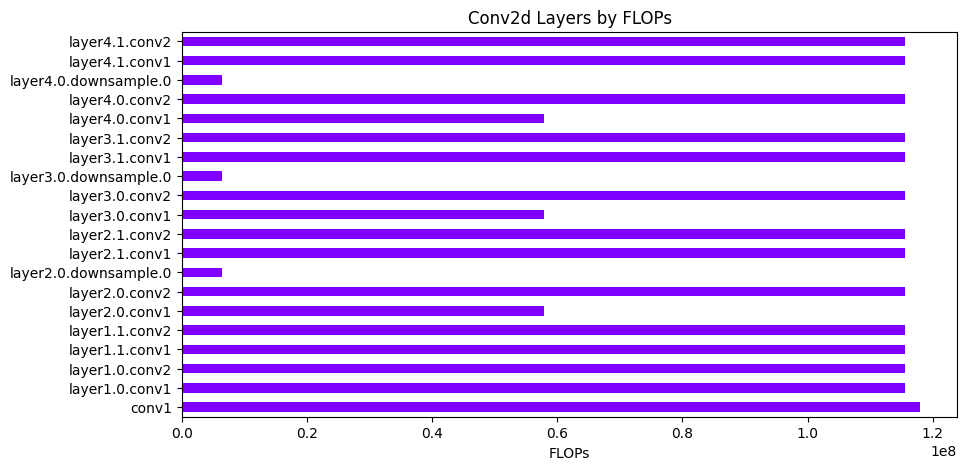
analyzer.find_layers_by_type("Conv2d", readable=True)
[11]:
| ProfilerMetric.NAME | ProfilerMetric.TYPE | ProfilerMetric.INPUTS | ProfilerMetric.OUTPUTS | ProfilerMetric.PARAMS | ProfilerMetric.PARAMS_TRAINABLE | ProfilerMetric.FLOPS | ProfilerMetric.MADDS | ProfilerMetric.MEM_READ | ProfilerMetric.MEM_WRITE | ProfilerMetric.RUNTIME | ProfilerMetric.RMS | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | conv1 | Conv2d | (3, 224, 224) | (64, 112, 112) | 9.41K | 9.41K | 118.01M | 235.23M | 159.94K | 802.82K | 0.0019 | tensor(0.1297) |
| 5 | layer1.0.conv1 | Conv2d | (64, 56, 56) | (64, 56, 56) | 36.86K | 36.86K | 115.61M | 231.01M | 237.57K | 200.7K | 0.0012 | tensor(0.0535) |
| 8 | layer1.0.conv2 | Conv2d | (64, 56, 56) | (64, 56, 56) | 36.86K | 36.86K | 115.61M | 231.01M | 237.57K | 200.7K | 0.001 | tensor(0.0452) |
| 10 | layer1.1.conv1 | Conv2d | (64, 56, 56) | (64, 56, 56) | 36.86K | 36.86K | 115.61M | 231.01M | 237.57K | 200.7K | 0.0009 | tensor(0.0509) |
| 13 | layer1.1.conv2 | Conv2d | (64, 56, 56) | (64, 56, 56) | 36.86K | 36.86K | 115.61M | 231.01M | 237.57K | 200.7K | 0.0011 | tensor(0.0440) |
| 15 | layer2.0.conv1 | Conv2d | (64, 56, 56) | (128, 28, 28) | 73.73K | 73.73K | 57.8M | 115.51M | 274.43K | 100.35K | 0.0006 | tensor(0.0416) |
| 18 | layer2.0.conv2 | Conv2d | (128, 28, 28) | (128, 28, 28) | 147.46K | 147.46K | 115.61M | 231.11M | 247.81K | 100.35K | 0.001 | tensor(0.0340) |
| 20 | layer2.0.downsample.0 | Conv2d | (64, 56, 56) | (128, 28, 28) | 8.19K | 8.19K | 6.42M | 12.74M | 208.9K | 100.35K | 0.00038 | tensor(0.0707) |
| 22 | layer2.1.conv1 | Conv2d | (128, 28, 28) | (128, 28, 28) | 147.46K | 147.46K | 115.61M | 231.11M | 247.81K | 100.35K | 0.0009 | tensor(0.0342) |
| 25 | layer2.1.conv2 | Conv2d | (128, 28, 28) | (128, 28, 28) | 147.46K | 147.46K | 115.61M | 231.11M | 247.81K | 100.35K | 0.001 | tensor(0.0301) |
| 27 | layer3.0.conv1 | Conv2d | (128, 28, 28) | (256, 14, 14) | 294.91K | 294.91K | 57.8M | 115.56M | 395.26K | 50.18K | 0.0007 | tensor(0.0291) |
| 30 | layer3.0.conv2 | Conv2d | (256, 14, 14) | (256, 14, 14) | 589.82K | 589.82K | 115.61M | 231.16M | 640.0K | 50.18K | 0.0011 | tensor(0.0251) |
| 32 | layer3.0.downsample.0 | Conv2d | (128, 28, 28) | (256, 14, 14) | 32.77K | 32.77K | 6.42M | 12.79M | 133.12K | 50.18K | 0.00032 | tensor(0.0330) |
| 34 | layer3.1.conv1 | Conv2d | (256, 14, 14) | (256, 14, 14) | 589.82K | 589.82K | 115.61M | 231.16M | 640.0K | 50.18K | 0.0012 | tensor(0.0224) |
| 37 | layer3.1.conv2 | Conv2d | (256, 14, 14) | (256, 14, 14) | 589.82K | 589.82K | 115.61M | 231.16M | 640.0K | 50.18K | 0.001 | tensor(0.0207) |
| 39 | layer4.0.conv1 | Conv2d | (256, 14, 14) | (512, 7, 7) | 1.18M | 1.18M | 57.8M | 115.58M | 1.23M | 25.09K | 0.0013 | tensor(0.0199) |
| 42 | layer4.0.conv2 | Conv2d | (512, 7, 7) | (512, 7, 7) | 2.36M | 2.36M | 115.61M | 231.19M | 2.38M | 25.09K | 0.0033 | tensor(0.0174) |
| 44 | layer4.0.downsample.0 | Conv2d | (256, 14, 14) | (512, 7, 7) | 131.07K | 131.07K | 6.42M | 12.82M | 181.25K | 25.09K | 0.0007 | tensor(0.0328) |
| 46 | layer4.1.conv1 | Conv2d | (512, 7, 7) | (512, 7, 7) | 2.36M | 2.36M | 115.61M | 231.19M | 2.38M | 25.09K | 0.0038 | tensor(0.0180) |
| 49 | layer4.1.conv2 | Conv2d | (512, 7, 7) | (512, 7, 7) | 2.36M | 2.36M | 115.61M | 231.19M | 2.38M | 25.09K | 0.0024 | tensor(0.0132) |
Analyze memory usage#
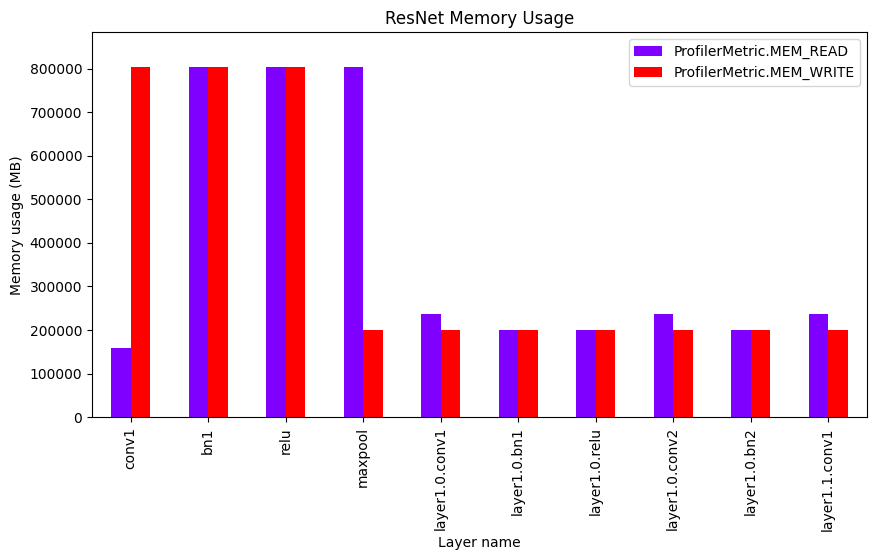
The analyze_memory_usage method can be used to analyze the memory usage of the model. It returns a pandas.DataFrame object with the memory usage of each layer.
[12]:
analyzer.analyze_memory_usage(readable=True).head(
5
) # only first 5 layers (for brevity)
[12]:
| ProfilerMetric.NAME | ProfilerMetric.MEM_READ | ProfilerMetric.MEM_WRITE | |
|---|---|---|---|
| 1 | conv1 | 159.94K | 802.82K |
| 2 | bn1 | 802.94K | 802.82K |
| 3 | relu | 802.82K | 802.82K |
| 4 | maxpool | 802.82K | 200.7K |
| 5 | layer1.0.conv1 | 237.57K | 200.7K |
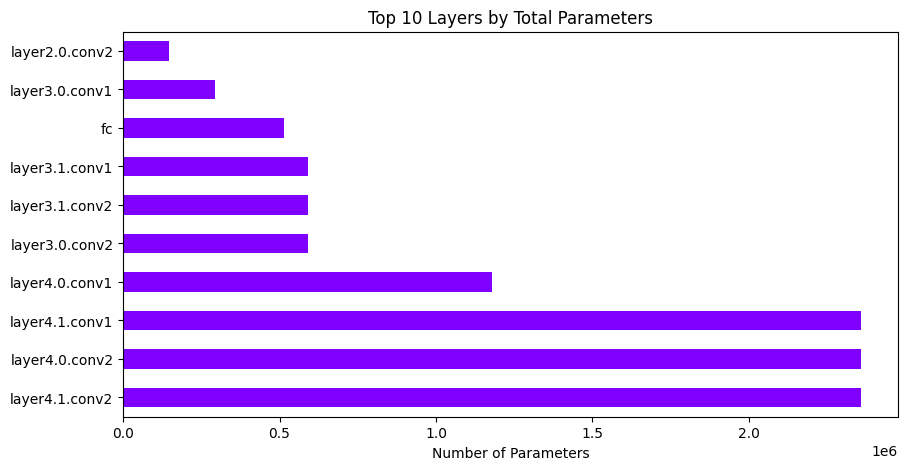
Plotting the results#
This section is under construction
[13]:
figsize = (10, 5)
analyzer.plot_top_n_layers(n=15, figsize=figsize, colormap="rainbow").show()
analyzer.plot_total_params_per_layer(
figsize=figsize, colormap="rainbow", limit=10
).show()
analyzer.plot_memory_usage(figsize=figsize, colormap="rainbow").show()
analyzer.plot_layer_wise_impact(
ProfilerMetric.RUNTIME, figsize=figsize, colormap="tab20"
).show()
analyzer.plot_layers_by_type(
"Conv2d", metric=ProfilerMetric.FLOPS, figsize=figsize, colormap="rainbow"
).show()